Torin 1
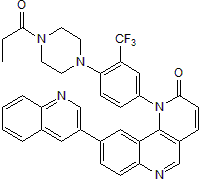
Chemical Name: 1-[4-[4-(1-Oxopropyl)-1-piperazinyl]-3-(trifluoromethyl)phenyl]-9-(3-quinolinyl)-benzo[h]-1,6-naphthyridin-2(1H)-one
Purity: ≥98%
Biological Activity
Torin 1 is a potent and selective ATP-competitive mTOR inhibitor (IC50 = 2 - 10 nM for mTORC1 and mTORC2). Torin 1 displays 200-fold selectivity for mTOR over DNA-PK, ATM and hVps34. In HeLa cells, Torin 1 induces autophagy. Torin 1 impairs cell growth and proliferation by suppression of the rapamycin-resistant functions of mTORC1. In human monocytes and myeloid dendritic cells, Torin 1 prevents decreases the anti-inflammatory potency of glucocorticoids. In the human endocrine cell line BON, Torin 1 increases neurotensin secretion and gene expression through MEK/ERK/c-Jun pathway activation.Technical Data
The technical data provided above is for guidance only.
For batch specific data refer to the Certificate of Analysis.
Tocris products are intended for laboratory research use only, unless stated otherwise.
Additional Information
Background References
-
An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1.
Thoreen CC, Kang SA, Chang JW et al.
J Biol Chem -
Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles.
Galluzzi et al.
Nat.Rev.Drug.Discov., 2017; -
The pharmacology of mTOR inhibition.
Guertin and Sabatini
Sci.Signal., 2009;2:pe24 -
mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway.
Peterson et al.
Cell., 2011;146:408 -
Discovery of 1-(4(-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benzo[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer.
Liu et al.
J.Med.Chem., 2010;53:7146
Product Datasheets
Reconstitution Calculator
Molarity Calculator
Citations for Torin 1
The citations listed below are publications that use Tocris products. Selected citations for Torin 1 include:
249 Citations: Showing 1 - 10
-
A central role for regulated protein stability in the control of TFE3 and MITF by nutrients.
Authors: Yumei Et al.
Mol Cell 2023;83:57-73.e9
-
TFEB-mediated lysosomal exocytosis alleviates high-fat diet-induced lipotoxicity in the kidney.
Authors: Atsushi Et al.
JCI Insight 2023;8
-
TFEB-dependent lysosome biogenesis is required for senescence.
Authors: Rachel Et al.
EMBO J 2023;42:e111241
-
Transcriptome, proteome, and protein synthesis within the intracellular cytomatrix.
Authors: Sandra L Et al.
iScience 2023;26:105965
-
Selective Eradication of Colon Cancer Cells Harboring PI3K and/or MAPK Pathway Mutations in 3D Culture by Combined PI3K/AKT/mTOR Pathway and MEK Inhibition.
Authors: Markus Et al.
Int J Mol Sci 2023;24
-
RagD auto-activating mutations impair MiT/TFE activity in kidney tubulopathy and cardiomyopathy syndrome.
Authors: Giancarlo Et al.
Nat Commun 2023;14:2775
-
Defects in lysosomal function and lipid metabolism in human microglia harboring a TREM2 loss of function mutation.
Authors: Wendy H Et al.
Acta Neuropathol 2023;145:749-772
-
Environmental control of Pub1 (NEDD4 family E3 ligase) in Schizosaccharomyces pombe is regulated by TORC2 and Gsk3.
Authors: Philip G Et al.
Life Sci Alliance 2022;5
-
LAMTOR1 inhibition of TRPML1-dependent lysosomal calcium release regulates dendritic lysosome trafficking and hippocampal neuronal function.
Authors: Yan Et al.
EMBO J 2022;41:e108119
-
Stress-induced perturbations in intracellular amino acids reprogram mRNA translation in osmoadaptation independently of the ISR.
Authors: Jing Et al.
Cell Rep 2022;40:111092
-
Reciprocal effects of mTOR inhibitors on pro-survival proteins dictate therapeutic responses in tuberous sclerosis complex.
Authors: Chin-Lee Et al.
iScience 2022;25:105458
-
Measuring Autophagic Cargo Flux with Keima-Based Probes.
Authors: Lisa B Et al.
Methods Mol Biol 2022;2445:99-115
-
Phosphorylation by casein kinase 2 enhances the interaction between ER-phagy receptor TEX264 and ATG8 proteins.
Authors: Koji L Et al.
EMBO Rep 2022;23:e54801
-
The FACT complex facilitates expression of lysosomal and antioxidant genes through binding to TFEB and TFE3.
Authors: Jose A Et al.
Autophagy 2022;18:2333-2349
-
SHISA5/SCOTIN restrains spontaneous autophagy induction by blocking contact between the ERES and phagophores.
Authors: Joo-Yeon Et al.
Autophagy 2022;18:1613-1628
-
Identification of a novel compound that simultaneously impairs the ubiquitin-proteasome system and autophagy.
Authors: Florian A Et al.
Autophagy 2022;18:1486-1502
-
DJ-1 is an essential downstream mediator in PINK1/parkin-dependent mitophagy.
Authors: Catherine Et al.
Brain 2022;145:4368-4384
-
Targeting of Ubiquitin E3 Ligase RNF5 as a Novel Therapeutic Strategy in Neuroectodermal Tumors.
Authors: Alessandra Et al.
Cancers (Basel) 2022;14
-
SETD2 transcriptional control of ATG14L/S isoforms regulates autophagosome-lysosome fusion.
Authors: Daniel J Et al.
Cell Death Dis 2022;13:953
-
mTORC1 controls Golgi architecture and vesicle secretion by phosphorylation of SCYL1.
Authors: Florian Et al.
Nat Commun 2022;13:4685
-
Compounds activating VCP D1 ATPase enhance both autophagic and proteasomal neurotoxic protein clearance.
Authors: Keith Et al.
Nat Commun 2022;13:4146
-
A novel ATG5 interaction with Ku70 potentiates DNA repair upon genotoxic stress.
Authors: Devrim Et al.
Sci Rep 2022;12:8134
-
Overexpressed Smurf1 is degraded in glioblastoma cells through autophagy in a p62-dependent manner.
Authors: Xinyi Et al.
FEBS Open Bio 2022;12:118-129
-
Novel Regulators of Macropinocytosis-Dependent Growth Revealed by Informer Set Library Screening in Pancreatic Cancer Cells.
Authors: Frank Et al.
Metabolites 2022;12
-
3D chromatin remodeling potentiates transcriptional programs driving cell invasion.
Authors: Adriana Et al.
Proc Natl Acad Sci U S A 2022;119:e2203452119
-
A pulse-chasable reporter processing assay for mammalian autophagic flux with HaloTag.
Authors: Noboru Et al.
Elife 2022;11
-
Local anesthetics elicit immune-dependent anticancer effects.
Authors: Peter Et al.
J Immunother Cancer 2022;10
-
Adaptive translational pausing is a hallmark of the cellular response to severe environmental stress.
Authors: Jing Et al.
Mol Cell 2021;81:4191-4208.e8
-
mTORC1 stimulates cell growth through SAM synthesis and m6A mRNA-dependent control of protein synthesis.
Authors: John M Et al.
Mol Cell 2021;81:2076-2093.e9
-
mTORC1 promotes cell growth via m6A-dependent mRNA degradation.
Authors: Joshua D Et al.
Mol Cell 2021;81:2064-2075.e8
-
Activation of mTORC1 at late endosomes misdirects T cell fate decision in older individuals.
Authors: Qiong Et al.
Sci Immunol 2021;6
-
p38 regulates the tumor suppressor PDCD4 via the TSC-mTORC1 pathway.
Authors: Markus Et al.
Cell Stress 2021;5:176-182
-
FTLD Patient-Derived Fibroblasts Show Defective Mitochondrial Function and Accumulation of p62.
Authors: Päivi Et al.
Mol Neurobiol 2021;58:5438-5458
-
Rheb mediates neuronal-activity-induced mitochondrial energetics through mTORC1-independent PDH activation.
Authors: Paul F Et al.
Dev Cell 2021;56:811-825.e6
-
NPC1-mTORC1 Signaling Couples Cholesterol Sensing to Organelle Homeostasis and Is a Targetable Pathway in Niemann-Pick Type C.
Authors: Roberto Et al.
Dev Cell 2021;56:260-276.e7
-
Glucose starvation induces autophagy via ULK1-mediated activation of PIKfyve in an AMPK-dependent manner.
Authors: David C Et al.
Dev Cell 2021;56:1961-1975.e5
-
Hepatic mTORC1 signaling activates ATF4 as part of its metabolic response to feeding and insulin.
Authors: John M Et al.
Mol Metab 2021;53:101309
-
Genotoxic stress in constitutive trisomies induces autophagy and the innate immune response via the cGAS-STING pathway.
Authors: Neysan Et al.
Commun Biol 2021;4:831
-
The mTOR regulated RNA-binding protein LARP1 requires PABPC1 for guided mRNA interaction.
Authors: Carolyn Et al.
Nucleic Acids Res 2021;49:458-478
-
mTORC1 promotes TOP mRNA translation through site-specific phosphorylation of LARP1.
Authors: An-Dao Et al.
Nucleic Acids Res 2021;49:3461-3489
-
Patient-specific iPSCs carrying an SFTPC mutation reveal the intrinsic alveolar epithelial dysfunction at the inception of interstitial lung disease.
Authors: Andrew Et al.
Cell Rep 2021;36:109636
-
Amino acids and mechanistic target of rapamycin regulate the fate of live engulfed cells.
Authors: Michael Et al.
FASEB J 2021;35:e21909
-
Increased fidelity of protein synthesis extends lifespan.
Authors: Mario Et al.
Cell Metab 2021;33:2288-2300.e12
-
Asparagine couples mitochondrial respiration to ATF4 activity and tumor growth.
Authors: Costas A Et al.
Cell Metab 2021;33:1013-1026.e6
-
pHLARE: a new biosensor reveals decreased lysosome pH in cancer cells.
Authors: Torsten Et al.
Mol Biol Cell 2021;32:131-142
-
Development of a specific live-cell assay for native autophagic flux.
Authors: Sami J Et al.
J Biol Chem 2021;297:101003
-
Oleate-induced aggregation of LC3 at the trans-Golgi network is linked to a protein trafficking blockade.
Authors: Guido Et al.
Cell Death Differ 2021;28:1733-1752
-
Rag GTPases and phosphatidylinositol 3-phosphate mediate recruitment of the AP-5/SPG11/SPG15 complex.
Authors: Anne-Claude Et al.
J Cell Biol 2021;220
-
Genistein Activates Transcription Factor EB and Corrects Niemann-Pick C Phenotype.
Authors: Juan Et al.
Int J Mol Sci 2021;22
-
Impaired TRIM16-Mediated Lysophagy in Chronic Obstructive Pulmonary Disease Pathogenesis.
Authors: Jun Et al.
J Immunol 2021;207:65-76
-
Comparing mTOR inhibitor Rapamycin with Torin-2 within the RIST molecular-targeted regimen in neuroblastoma cells.
Authors: Selim Et al.
Int J Med Sci 2021;18:137-149
-
TMBIM6 (transmembrane BAX inhibitor motif containing 6) enhances autophagy through regulation of lysosomal calcium.
Authors: Sung Hoon Et al.
Autophagy 2021;17:761-778
-
Trehalose causes low-grade lysosomal stress to activate TFEB and the autophagy-lysosome biogenesis response.
Authors: Joel D Et al.
Autophagy 2021;17:3740-3752
-
A high-throughput screening identifies ZNF418 as a novel regulator of the ubiquitin-proteasome system and autophagy-lysosomal pathway.
Authors: Hanna Et al.
Autophagy 2021;17:3124-3139
-
The DNA methyltransferase DNMT3A contributes to autophagy long-term memory.
Authors: Michael G Et al.
Autophagy 2021;17:1259-1277
-
Generation of mouse-human chimeric embryos.
Authors: Boyang Et al.
Nat Protoc 2021;16:3954-3980
-
Atg7 deficiency in microglia drives an altered transcriptomic profile associated with an impaired neuroinflammatory response.
Authors: Mathilde Et al.
Mol Brain 2021;14:87
-
Evidence for Lysosomal Dysfunction within the Epidermis in Psoriasis and Atopic Dermatitis.
Authors: Hans-Uwe Et al.
J Invest Dermatol 2021;141:2838-2848.e4
-
Adipocytes disrupt the translational programme of acute lymphoblastic leukaemia to favour tumour survival and persistence.
Authors: M Et al.
Nat Commun 2021;12:5507
-
Loss of SNORA73 reprograms cellular metabolism and protects against steatohepatitis.
Authors: Shuang Et al.
Nat Commun 2021;12:5214
-
Alt-RPL36 downregulates the PI3K-AKT-mTOR signaling pathway by interacting with TMEM24.
Authors: Yang Et al.
Nat Commun 2021;12:508
-
NEK9 regulates primary cilia formation by acting as a selective autophagy adaptor for MYH9/myosin IIA.
Authors: Koji L Et al.
Nat Commun 2021;12:3292
-
Cdk5 and GSK3β inhibit fast endophilin-mediated endocytosis.
Authors: Konstantinos Et al.
Nat Commun 2021;12:2424
-
The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals.
Authors: John M Et al.
Elife 2021;10
-
3D Culture Models with CRISPR Screens Reveal Hyperactive NRF2 as a Prerequisite for Spheroid Formation via Regulation of Proliferation and Ferroptosis.
Authors: Steven P Et al.
Mol Cell 2020;80:828-844.e6
-
Transient inhibition of mTOR in human pluripotent stem cells enables robust formation of mouse-human chimeric embryos.
Authors: Jingxin Et al.
Sci Adv 2020;6:eaaz0298
-
A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome.
Authors: Pier Paolo Et al.
Nature 2020;585:597-602
-
Decoy exosomes provide protection against bacterial toxins.
Authors: Derya Et al.
Nature 2020;579:260-264
-
Microglia promote glioblastoma via mTOR-mediated immunosuppression of the tumour microenvironment.
Authors: Jun Et al.
EMBO J 2020;39:e103790
-
mTORC2 Assembly Is Regulated by USP9X-Mediated Deubiquitination of RICTOR.
Authors: David C Et al.
Cell Rep 2020;33:108564
-
CDK8 Fine-Tunes IL-6 Transcriptional Activities by Limiting STAT3 Resident Time at the Gene Loci.
Authors: Ignacio Et al.
Cell Rep 2020;33:108545
-
Protein Stability Buffers the Cost of Translation Attenuation following eIF2α Phosphorylation.
Authors: Anne Et al.
Cell Rep 2020;32:108154
-
BCR-Induced Ca2+ Signals Dynamically Tune Survival, Metabolic Reprogramming, and Proliferation of Naive B Cells.
Authors: Christopher J Et al.
Cell Rep 2020;31:107474
-
The GATA Transcription Factor Gaf1 Represses tRNAs, Inhibits Growth, and Extends Chronological Lifespan Downstream of Fission Yeast TORC1.
Authors: Jürg Et al.
Cell Rep 2020;30:3240-3249.e4
-
Macropinocytosis Renders a Subset of Pancreatic Tumor Cells Resistant to mTOR Inhibition.
Authors: Jurre J Et al.
Cell Rep 2020;30:2729-2742.e4
-
Von Hippel-Lindau tumor suppressor (VHL) stimulates TOR signaling by interacting with phosphoinositide 3-kinase (PI3K).
Authors: Wonho Et al.
J Biol Chem 2020;295:2336-2347
-
The intraflagellar transport protein IFT20 controls lysosome biogenesis by regulating the post-Golgi transport of acid hydrolases.
Authors: Cosima T Et al.
Cell Death Differ 2020;27:310-328
-
Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Authors: Satoru Et al.
Nat Struct Mol Biol 2020;27:1194-1201
-
LATS suppresses mTORC1 activity to directly coordinate Hippo and mTORC1 pathways in growth control.
Authors: Chen Et al.
Nat Cell Biol 2020;22:246-256
-
Effects of mTOR inhibitors on neuropathic pain revealed by optical imaging of the insular cortex in rats.
Authors: Bae Hwan Et al.
Brain Res 2020;1733:146720
-
Oxidation of multiple MiT/TFE transcription factors links oxidative stress to transcriptional control of autophagy and lysosome biogenesis.
Authors: Yan Et al.
Autophagy 2020;16:1683-1696
-
De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy.
Authors: Vladimir Et al.
Autophagy 2020;16:1044-1060
-
MITF and TFEB cross-regulation in melanoma cells.
Authors: Valerie Et al.
PLoS One 2020;15:e0238546
-
The small GTPase Rab32 resides on lysosomes to regulate mTORC1 signaling.
Authors: Jing Et al.
J Cell Sci 2020;133
-
Location-specific inhibition of Akt reveals regulation of mTORC1 activity in the nucleus.
Authors: JoAnn Et al.
Nat Commun 2020;11:6088
-
Cathepsin D deficiency in mammary epithelium transiently stalls breast cancer by interference with mTORC1 signaling.
Authors: Florian Et al.
Nat Commun 2020;11:5133
-
Phosphorylation of eukaryotic initiation factor-2α (eIF2α) in autophagy.
Authors: Guido Et al.
Cell Death Dis 2020;11:433
-
Macropinocytosis drives T cell growth by sustaining the activation of mTORC1.
Authors: Di Et al.
Nat Commun 2020;11:180
-
Microglial metabolic flexibility supports immune surveillance of the brain parenchyma.
Authors: Louis-Philippe Et al.
Nat Commun 2020;11:1559
-
Rap1-GTPases control mTORC1 activity by coordinating lysosome organization with amino acid availability.
Authors: Sang Gyun Et al.
Nat Commun 2020;11:1416
-
Resolvin E1 is a pro-repair molecule that promotes intestinal epithelial wound healing.
Authors: Asma Et al.
Proc Natl Acad Sci U S A 2020;117:9477-9482
-
The small GTPase Rab32 resides on lysosomes to regulate mTORC1 signaling.
Authors: Jing Et al.
J Cell Sci 2020;
-
HIF1A and NFAT5 coordinate Na+-boosted antibacterial defense via enhanced autophagy and autolysosomal targeting.
Authors: Neubert Et al.
Autophagy 2019;:1
-
Lysophosphatidic acid represses autophagy in prostate carcinoma cells.
Authors: Roberto J Et al.
Biochem Cell Biol 2019;97:387-396
-
Mammalian cell growth dynamics in mitosis.
Authors: Scott R Et al.
Elife 2019;8
-
Raptor-Mediated Proteasomal Degradation of Deamidated 4E-BP2 Regulates Postnatal Neuronal Translation and NF-κB Activity.
Authors: Seyed Mehdi Et al.
Cell Rep 2019;29:3620-3635.e7
-
The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress.
Authors: Andreas Et al.
J Biol Chem 2019;294:8197-8217
-
NOX1-Dependent mTORC1 Activation via S100A9 Oxidation in Cancer Stem-like Cells Leads to Colon Cancer Progression.
Authors: Masaya Et al.
Cell Rep 2019;28:1282-1295.e8
-
Lethal Poisoning of Cancer Cells by Respiratory Chain Inhibition plus Dimethyl α-Ketoglutarate.
Authors: Sica Et al.
Cell Rep 2019;27:820
-
Type 2 diabetes is associated with suppression of autophagy and lipid accumulation in β-cells.
Authors: Ji Et al.
J Cell Mol Med 2019;23:2890
-
ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C.
Authors: Daniel S Et al.
Nat Cell Biol 2019;21:1206-1218
-
Involvement of Lamin B1 Reduction in Accelerated Cellular Senescence during Chronic Obstructive Pulmonary Disease Pathogenesis.
Authors: Jun Et al.
J Immunol 2019;202:1428-1440
-
Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition.
Authors: Charles A Et al.
Nat Chem Biol 2019;15:776-785
-
MITF-MIR211 axis is a novel autophagy amplifier system during cellular stress.
Authors: Arzu Et al.
Autophagy 2019;15:375-390
-
Impaired TFEB-mediated lysosomal biogenesis promotes the development of pancreatitis in mice and is associated with human pancreatitis.
Authors: Hua Et al.
Autophagy 2019;15:1954-1969
-
TFEB-driven endocytosis coordinates MTORC1 signaling and autophagy.
Authors: Andrea Et al.
Autophagy 2019;15:151-164
-
VCP maintains lysosomal homeostasis and TFEB activity in differentiated skeletal muscle.
Authors: Conrad C Et al.
Autophagy 2019;15:1082-1099
-
Primary cilia mediate mitochondrial stress responses to promote dopamine neuron survival in a Parkinson's disease model.
Authors: Dong-Hyung Et al.
Cell Death Dis 2019;10:952
-
Dysfunctional autophagy following exposure to pro-inflammatory cytokines contributes to pancreatic β-cell apoptosis.
Authors: Lambelet Et al.
Cell Death Dis 2018;9:96
-
Amino acids stimulate the endosome-to-Golgi trafficking through Ragulator and small GTPase Arl5.
Authors: Shi Et al.
Nat Commun 2018;9:4987
-
Is Ras a potential target in treatment against cutaneous squamous cell carcinoma?.
Authors: Li Et al.
J Cancer 2018;9:3373
-
mTOR-dependent phosphorylation controls TFEB nuclear export.
Authors: Napolitano Et al.
Nat Commun 2018;9:3312
-
Carbon monoxide-induced TFEB nuclear translocation enhances mitophagy/mitochondrial biogenesis in hepatocytes and ameliorates inflammatory liver injury.
Authors: Kim Et al.
Cell Death Dis 2018;9:1060
-
The Long-lived Protein Degradation Assay: an Efficient Method for Quantitative Determination of the Autophagic Flux of Endogenous Proteins in Adherent Cell Lines.
Authors: Nikolai Et al.
Bio Protoc 2018;8:e2836
-
mTORC1 Promotes Metabolic Reprogramming by the Suppression of GSK3-Dependent Foxk1 Phosphorylation.
Authors: Xin Et al.
Mol Cell 2018;70:949-960.e4
-
Phosphatidylinositol-5-Phosphate 4-Kinases Regulate Cellular Lipid Metabolism By Facilitating Autophagy.
Authors: Lundquist Et al.
Mol Cell 2018;70:531
-
PTBP1-Mediated Alternative Splicing Regulates the Inflammatory Secretome and the Pro-tumorigenic Effects of Senescent Cells.
Authors: Georgilis Et al.
Cancer Cell 2018;34:85
-
Cancer Cells Co-opt the Neuronal Redox-Sensing Channel TRPA1 to Promote Oxidative-Stress Tolerance.
Authors: Yasuo Et al.
Cancer Cell 2018;33:985-1003.e7
-
Metabolic reprogramming of murine cardiomyocytes during autophagy requires the extracellular nutrient sensor decorin.
Authors: Gubbiotti Et al.
J Biol Chem 2018;293:16940
-
Translational and HIF-1α-Dependent Metabolic Reprogramming Underpin Metabolic Plasticity and Responses to Kinase Inhibitors and Biguanides.
Authors: Peter Et al.
Cell Metab 2018;28:817-832.e8
-
ULK1 O-GlcNAcylation Is Crucial for Activating VPS34 via ATG14L during Autophagy Initiation.
Authors: Pyo Et al.
Cell Rep 2018;25:2878
-
Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17.
Authors: Matsui Et al.
J Cell Biol 2018;217:2633
-
Lysosomal Protein Lamtor1 Controls Innate Immune Responses via Nuclear Translocation of Transcription Factor EB.
Authors: Hayama Et al.
J Immunol 2018;200:3790
-
Integration of Distinct ShcA Signaling Complexes Promotes Breast Tumor Growth and Tyrosine Kinase Inhibitor Resistance.
Authors: Ha Et al.
Mol Cancer Res 2018;16:894
-
HEPES activates a MiT/TFE-dependent lysosomal-autophagic gene network in cultured cells: A call for caution.
Authors: Tol Et al.
Autophagy 2018;14:437
-
The pro-oxidant adaptor p66SHC promotes B cell mitophagy by disrupting mitochondrial integrity and recruiting LC3-II.
Authors: Cosima T Et al.
Autophagy 2018;14:2117-2138
-
Role of the MAPK/cJun NH2-terminal kinase signaling pathway in starvation-induced autophagy.
Authors: Barutcu Et al.
Autophagy 2018;14:1586
-
Lifespan extension without fertility reduction following dietary addition of the autophagy activator Torin1 in Drosophila melanogaster.
Authors: Mason Et al.
PLoS One 2018;13:e0190105
-
Mapping the mammalian ribosome quality control complex interactome using proximity labeling approaches.
Authors: Zuzow Et al.
Mol Biol Cell 2018;
-
Tenovin-6 impairs autophagy by inhibiting autophagic flux.
Authors: Yuan Et al.
Cell Death Dis 2017;8:e2608
-
SYK kinase mediates brown fat differentiation and activation.
Authors: Lei Et al.
Nat Commun 2017;8:2115
-
Primary fibroblasts from CSPα mutation carriers recapitulate hallmarks of the adult onset neuronal ceroid lipofuscinosis.
Authors: Mark S Et al.
Sci Rep 2017;7:6332
-
TBK1 Provides Context-Selective Support of the Activated AKT/mTOR Pathway in Lung Cancer.
Authors: Cooper Et al.
Cancer Res 2017;77:5077
-
Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway.
Authors: Guerroue Et al.
Mol Cell 2017;68:786
-
m6A Facilitates eIF4F-Independent mRNA Translation.
Authors: Jun Et al.
Mol Cell 2017;68:504-514.e7
-
mTORC1 Phosphorylates Acetyltransferase p300 to Regulate Autophagy and Lipogenesis.
Authors: Li Et al.
Mol Cell 2017;68:323-335.e6
-
mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1.
Authors: Heidi M Et al.
Mol Cell 2017;67:922-935.e5
-
PARK2 Depletion Connects Energy and Oxidative Stress to PI3K/Akt Activation via PTEN S-Nitrosylation.
Authors: Gupta Et al.
Mol Cell 2017;65:999
-
Akt regulation of glycolysis mediates bioenergetic stability in epithelial cells.
Authors: Hung Et al.
Elife 2017;6
-
mTORC1-Mediated Inhibition of 4EBP1 Is Essential for Hedgehog Signaling-Driven Translation and Medulloblastoma.
Authors: Yong Ha Et al.
Dev Cell 2017;43:673-688.e5
-
DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy.
Authors: Nguyen Et al.
Dev Cell 2017;42:9
-
PtdIns3P controls mTORC1 signaling through lysosomal positioning.
Authors: Hong Et al.
J Cell Biol 2017;216:4217
-
Tim-3 is a Marker of Plasmacytoid Dendritic Cell Dysfunction during HIV Infection and Is Associated with the Recruitment of IRF7 and p85 into Lysosomes and with the Submembrane Displacement of TLR9.
Authors: Schwartz Et al.
J Immunol 2017;198:3181
-
TORC1 and TORC2 converge to regulate the SAGA co-activator in response to nutrient availability.
Authors: Laboucarié Et al.
EMBO Rep 2017;18:2197
-
Deubiquitinase OTUD6B Isoforms Are Important Regulators of Growth and Proliferation.
Authors: Sobol Et al.
Mol Cancer Res 2017;15:117
-
Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication.
Authors: Michael S Et al.
PLoS Pathog 2017;13:e1006635
-
The E3 ubiquitin ligase NEDD4 enhances killing of membrane-perturbing intracellular bacteria by promoting autophagy.
Authors: Pei Et al.
Autophagy 2017;13:2041
-
Loss of ULK1 increases RPS6KB1-NCOR1 repression of NR1H/LXR-mediated Scd1 transcription and augments lipotoxicity in hepatic cells.
Authors: Sinha Et al.
Autophagy 2017;13:169
-
Nonradioactive quantification of autophagic protein degradation with L-azidohomoalanine labeling.
Authors: Qingsong Et al.
Nat Protoc 2017;12:279-288
-
Tracking the Activity of mTORC1 in Living Cells Using Genetically Encoded FRET-based Biosensor TORCAR.
Authors: Jin Et al.
Curr Protoc Chem Biol 2016;8:225-233
-
The anti-tumor drug 2-hydroxyoleic acid (Minerval) stimulates signaling and retrograde transport.
Authors: Torgersen Et al.
Oncotarget 2016;7:86871
-
TSC loss distorts DNA replication programme and sensitises cells to genotoxic stress.
Authors: Pai Et al.
Oncotarget 2016;7:85365
-
Diagnostic and clinical relevance of the autophago-lysosomal network in human gliomas.
Authors: Jennewein Et al.
Oncotarget 2016;7:20016
-
Polarization of M2 macrophages requires Lamtor1 that integrates cytokine and amino-acid signals
Authors: Kimura Et al.
Nature Communications 2016;7:13130
-
mTORC1 and CK2 coordinate ternary and eIF4F complex assembly.
Authors: Gandin Et al.
Nat Commun 2016;7:11127
-
mTORC1-mediated inhibition of polycystin-1 expression drives renal cyst formation in tuberous sclerosis complex.
Authors: Pema Et al.
Mol Biol Cell 2016;7:10786
-
Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mTOR-independent pathway.
Authors: Chen Et al.
PLoS One 2016;6:28423
-
LKB1 loss links serine metabolism to DNA methylation and tumorigenesis.
Authors: Kottakis Et al.
Nature 2016;539:390
-
An evolutionarily conserved mechanism for cAMP elicited axonal regeneration involves direct activation of the dual leucine zipper kinase DLK.
Authors: Hao Et al.
Proc Natl Acad Sci U S A 2016;5
-
Characterizing inactive ribosomes in translational profiling.
Authors: Liu
Translation (Austin) 2016;4:e1138018
-
Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP.
Authors: Ricoult Et al.
Oncogene 2016;35:1250
-
TDP-43 loss of function increases TFEB activity and blocks autophagosome-lysosome fusion.
Authors: Xia Et al.
EMBO J 2016;35:121
-
A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets.
Authors: Li Et al.
Sci Adv 2016;2:e1601470
-
Nutrient-regulated Phosphorylation of ATG13 Inhibits Starvation-induced Autophagy.
Authors: Puente Et al.
J Biol Chem 2016;291:6026
-
RACK1 Is an Interaction Partner of ATG5 and a Novel Regulator of Autophagy.
Authors: Erbil Et al.
J Biol Chem 2016;291:16753
-
Human Cells Cultured under Physiological Oxygen Utilize Two Cap-binding Proteins to recruit Distinct mRNAs for Translation.
Authors: James Et al.
J Biol Chem 2016;291:10772-82
-
mTOR controls lysosome tubulation and antigen presentation in macrophages and dendritic cells.
Authors: Saric Et al.
Autophagy 2016;27:321
-
nanoCAGE reveals 5' UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs.
Authors: Gandin Et al.
Genome Res 2016;26:636
-
Involvement of PARK2-Mediated Mitophagy in Idiopathic Pulmonary Fibrosis Pathogenesis.
Authors: Kobayashi Et al.
J Immunol 2016;197:504
-
Neuronal gene repression in Niemann-Pick type C models is mediated by the c-Abl/HDAC2 signaling pathway.
Authors: Contreras Et al.
Biochim Biophys Acta 2016;1859:269
-
A Founder Mutation in VPS11 Causes an Autosomal Recessive Leukoencephalopathy Linked to Autophagic Defects.
Authors: Zhang Et al.
Cell Rep 2016;12:e1005848
-
Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway.
Authors: Bouche Et al.
Autophagy 2016;12:484
-
Hypoxia promotes noncanonical autophagy in nucleus pulposus cells independent of MTOR and HIF1A signaling.
Authors: Choi Et al.
Autophagy 2016;12:1631
-
Iron overload causes endolysosomal deficits modulated by NAADP-regulated 2-pore channels and RAB7A.
Authors: Fernández Et al.
Autophagy 2016;12:1487
-
TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages.
Authors: Pastore Et al.
Autophagy 2016;12:1240
-
TP53INP2/DOR, a mediator of cell autophagy, promotes rDNA transcription via facilitating the assembly of the POLR1/RNA polymerase I preinitiation complex at rDNA promoters.
Authors: Xu Et al.
Autophagy 2016;12:1118
-
IL-15 activates mTOR and primes stress-activated gene expression leading to prolonged antitumor capacity of NK cells.
Authors: Mao Et al.
Blood 2016;128:1475
-
IBMPFD Disease-Causing Mutant VCP/p97 Proteins Are Targets of Autophagic-Lysosomal Degradation.
Authors: Bayraktar Et al.
PLoS One 2016;11:e0164864
-
The Loss of Lam2 and Npr2-Npr3 Diminishes the Vacuolar Localization of Gtr1-Gtr2 and Disinhibits TORC1 Activity in Fission Yeast.
Authors: Ma Et al.
PLoS One 2016;11:e0156239
-
Trehalose upregulates progranulin expression in human and mouse models of GRN haploinsufficiency: a novel therapeutic lead to treat frontotemporal dementia.
Authors: Holler Et al.
Mol Neurodegener 2016;11:46
-
A Small Molecule Screen Exposes mTOR Signaling Pathway Involvement in Radiation-Induced Apoptosis.
Authors: John S Et al.
ACS Chem Biol 2016;11:1428-37
-
Activation of 5-HT7 receptor stimulates neurite elongation through mTOR, Cdc42 and actin filaments dynamics.
Authors: Speranza Et al.
Nat Chem Biol 2015;9:62
-
The MAPK and PI3K pathways mediate CNTF-induced neuronal survival and process outgrowth in hypothalamic organotypic cultures.
Authors: Askvig and Watt
J Cell Commun Signal 2015;9:217
-
The mTORC1/4E-BP pathway coordinates hemoglobin production with L-leucine availability.
Authors: Chung Et al.
Mol Cancer Res 2015;8:ra34
-
Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes.
Authors: Sagnier Et al.
J Virol 2015;89:615
-
Differential Phosphatidylinositol-3-Kinase-Akt-mTOR Activation by Semliki Forest and Chikungunya Viruses Is Dependent on nsP3 and Connected to Replication Complex Internalization.
Authors: Giuseppe Et al.
J Virol 2015;89:11420-37
-
Reinstating aberrant mTORC1 activity in Huntington's disease mice improves disease phenotypes.
Authors: Lee Et al.
Neuron 2015;85:303
-
Autophagy regulates hepatocyte identity and epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions promoting Snail degradation.
Authors: Grassi Et al.
Cell Death Dis 2015;6:e1880
-
Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A.
Authors: Wong Et al.
Nat Commun 2015;6:8048
-
Autophagy activation and protection from mitochondrial dysfunction in human chondrocytes.
Authors: Figueroa Et al.
Autophagy 2015;67:966
-
SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1.
Authors: Rebsamen Et al.
Autophagy 2015;519:477
-
Diacylglycerol kinase-ζ regulates mTORC1 and lipogenic metabolism in cancer cells through SREBP-1.
Authors: Torres-Ayuso Et al.
Oncogenesis 2015;4:e164
-
Decorin is an autophagy-inducible proteoglycan and is required for proper in vivo autophagy.
Authors: Gubbiotti Et al.
Matrix Biol 2015;48:14
-
Regulation of the transcription factor EB-PGC1α axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress.
Authors: Ma Et al.
Mol Cell Biol 2015;35:956
-
Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9.
Authors: Jung Et al.
Mol Cell Biol 2015;35:2479
-
HIV-1 Tat alters neuronal autophagy by modulating autophagosome fusion to the lysosome: implications for HIV-associated neurocognitive disorders.
Authors: Fields Et al.
J Neurosci 2015;35:1921
-
Activated α2-macroglobulin binding to human prostate cancer cells triggers insulin-like responses.
Authors: Uma Kant Et al.
J Biol Chem 2015;290:9571-87
-
La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1).
Authors: Fonseca Et al.
J Biol Chem 2015;290:15996
-
Cholesterol-mediated activation of acid sphingomyelinase disrupts autophagy in the retinal pigment epithelium.
Authors: Toops Et al.
J Exp Med 2015;26:41640
-
Amyloid precursor protein (APP) affects global protein synthesis in dividing human cells.
Authors: Sobol Et al.
J Cell Physiol 2015;230:1064
-
PDK1 orchestrates early NK cell development through induction of E4BP4 expression and maintenance of IL-15 responsiveness.
Authors: Yang Et al.
J Clin Invest 2015;212:253
-
Apoptotic effects of high-dose rapamycin occur in S-phase of the cell cycle.
Authors: Saqcena Et al.
Autophagy 2015;14:2285
-
Functional Proteomics Identifies Acinus L as a Direct Insulin- and Amino Acid-Dependent Mammalian Target of Rapamycin Complex 1 (mTORC1) Substrate.
Authors: Jörn Et al.
Mol Cell Proteomics 2015;14:2042-55
-
A mechanism for asymmetric cell division resulting in proliferative asynchronicity.
Authors: Dey-Guha Et al.
Autophagy 2015;13:223
-
mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation.
Authors: Moon Et al.
Cell Rep 2015;12:102
-
Coactivator SRC-2-dependent metabolic reprogramming mediates prostate cancer survival and metastasis.
Authors: Dasgupta Et al.
Sci Signal 2015;125:1174
-
Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication.
Authors: Avey Et al.
PLoS Pathog 2015;11:e1004993
-
GSK-3 modulates cellular responses to a broad spectrum of kinase inhibitors.
Authors: Thorne Et al.
Mol Biol Cell 2015;11:58
-
PARK2-mediated mitophagy is involved in regulation of HBEC senescence in COPD pathogenesis.
Authors: Ito Et al.
Nature 2015;11:547
-
Pharmacological inhibition of lysosomes activates the MTORC1 signaling pathway in chondrocytes in an autophagy-independent manner.
Authors: Newton Et al.
Autophagy 2015;11:1594
-
mTORC2 regulates cardiac response to stress by inhibiting MST1.
Authors: Sciarretta Et al.
Elife 2015;11:125
-
TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy.
Authors: Matsuzawa Et al.
Sci Rep 2015;11:1052
-
Hedgehog signaling activates a positive feedback mechanism involving Ins-like growth factors to induce osteoblast differentiation.
Authors: Shi Et al.
Front Behav Neurosci 2015;112:4678
-
Tor Signaling Regulates Transcription of Amino Acid Permeases through a GATA Transcription Factor Gaf1 in Fission Yeast.
Authors: Ma Et al.
PLoS One 2015;10:e0144677
-
Rapamycin-insensitive up-regulation of adipocyte phospholipase A2 in tuberous sclerosis and lymphangioleiomyomatosis.
Authors: Li Et al.
J Exp Med 2014;9:e104809
-
Fertilization-induced autophagy in mouse embryos is independent of mTORC1.
Authors: Yamamoto Et al.
Biol Reprod 2014;91:7
-
The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris.
Authors: Martina Et al.
Sci Signal 2014;7:ra9
-
Oncogenic mutations in adenomatous polyposis coli (Apc) activate mechanistic target of rapamycin complex 1 (mTORC1) in mice and zebrafish.
Authors: Valvezan Et al.
Dis Model Mech 2014;7:63
-
Palmitate induces mRNA translation and increases ER protein load in islet β-cells via activation of the mammalian target of rapamycin pathway.
Authors: Hatanaka Et al.
Diabetes 2014;63:3404
-
Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex.
Authors: Liang Et al.
J Exp Med 2014;211:2249
-
OE and mTORC2 cooperate to enhance prostaglandin biosynthesis and tumorigenesis in TSC2-deficient LAM cells.
Authors: Li Et al.
BMC Biochem 2014;211:15
-
Mammalian target of Rapamycin inhibition and mycobacterial survival are uncoupled in murine macrophages.
Authors: Zullo Et al.
Proc Natl Acad Sci U S A 2014;15:4
-
An invertebrate Warburg effect: a shrimp virus achieves successful replication by altering the host metabolome via the PI3K-Akt-mTOR pathway.
Authors: Su Et al.
PLoS Pathog 2014;10:e1004196
-
Dynamic modelling of pathways to cellular senescence reveals strategies for targeted interventions.
Authors: Viktor I Et al.
PLoS Comput Biol 2014;10:e1003728
-
Development of a novel method for quantification of autophagic protein degradation by AHA labeling.
Authors: Zhang Et al.
PLoS Genet 2014;10:901
-
Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids.
Authors: Sahani Et al.
Autophagy 2014;10:431
-
Autophagy fosters myofibroblast differentiation through MTORC2 activation and downstream upregulation of CTGF.
Authors: Bernard Et al.
Cell Cycle 2014;10:2193
-
Japanese encephalitis virus replication is negatively regulated by autophagy and occurs on LC3-I- and EDEM1-containing membranes.
Authors: Sharma Et al.
Arthritis Rheumatol 2014;10:1637
-
p53-directed translational control can shape and expand the universe of p53 target genes.
Authors: Zaccara Et al.
Cell Death Differ 2014;
-
Live imaging and single-cell analysis reveal differential dynamics of autophagy and apoptosis.
Authors: Xu Et al.
Autophagy 2013;9:1418
-
ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change.
Authors: Montané and Menand
Nat Commun 2013;64:4361
-
Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes.
Authors: Bachar-Wikstrom Et al.
Diabetes 2013;62:1227
-
Differential regulation of the expressions of the PGC-1α splice variants, lipins, and PPARα in heart compared to liver.
Authors: Kok Et al.
J Lipid Res 2013;54:1662
-
Selective targeting of human colon cancer stem-like cells by the mTOR inhibitor Torin-1.
Authors: Francipane and Lagasse
Oncotarget 2013;4:1948
-
Epigenetic regulation of autophagy by the methyltransferase G9a.
Authors: Narvajas Et al.
Mol Cell Biol 2013;33:3983
-
Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation.
Authors: Fournier Et al.
Mol Cell Biol 2013;33:2285
-
Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity.
Authors: Li Et al.
J Biol Chem 2013;288:35769
-
mTOR complex 2 phosphorylates IMP1 cotranslationally to promote IGF2 production and the proliferation of mouse embryonic fibroblasts.
Authors: Dai Et al.
Genes Dev 2013;27:301
-
SYK regulates mTOR signaling in AML.
Authors: Carnevale Et al.
Leukemia 2013;27:2118
-
mTOR regulates phagosome and entotic vacuole fission.
Authors: Krajcovic Et al.
Mol Biol Cell 2013;24:3736
-
Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion.
Authors: Zhou Et al.
Cell Res 2013;23:508
-
Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy.
Authors: Gammoh Et al.
Nat Struct Mol Biol 2013;20:144
-
Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes.
Authors: Martina and Puertollano
J Cell Biol 2013;200:475
-
mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation.
Authors: Morita Et al.
Cell Metab 2013;18:698
-
WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation.
Authors: Esen Et al.
Cell Metab 2013;17:745
-
Tumor VEGF:VEGFR2 autocrine feed-forward loop triggers angiogenesis in lung cancer.
Authors: Rolf A Et al.
J Clin Invest 2013;123:1732-40
-
MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB.
Authors: Martina Et al.
Autophagy 2012;8:903
-
Structure-activity analysis of niclosamide reveals potential role for cytoplasmic pH in control of mammalian target of rapamycin complex 1 (mTORC1) signaling.
Authors: Fonseca Et al.
J Biol Chem 2012;287:17530
-
Keap1 degradation by autophagy for the maintenance of redox homeostasis.
Authors: Taguchi Et al.
J Exp Bot 2012;109:13561
FAQs
No product specific FAQs exist for this product, however you may
View all Small Molecule FAQsReviews for Torin 1
There are currently no reviews for this product. Be the first to review Torin 1 and earn rewards!
Have you used Torin 1?
Submit a review and receive an Amazon gift card.
$25/€18/£15/$25CAN/¥75 Yuan/¥2500 Yen for a review with an image
$10/€7/£6/$10 CAD/¥70 Yuan/¥1110 Yen for a review without an image